UMD-USHbases -
Access to the databases UMD [1] is a generic software that allows the creation of Locus Specific DataBases. Using this tool, a set of databases has been developed in order to centralise and analyse data for the 7 most commonly involved genes: MYO7A, USH1C, CDH23, PCDH15 and USH1G for Usher type I, USH2A and USH3A for Usher type II and III respectively. You can enter one database by clicking on one of the link below:
|
This set is curated by the Usher
group from Montpellier.
When referring to UMD-USHbases, please cite:
Baux D, Faugère V, Larrieu L, Le Guédard-Méreuze S, Hamroun D, Béroud C, Malcolm S, Claustres M, Roux AF.
UMD-USHbases: a comprehensive set of databases to record and analyse pathogenic mutations and unclassified variants in seven Usher syndrome causing genes. Hum Mutat. 2008 May 16.
This software and data are provided to enhance knowledge and encourage progress in the scientific community and are to be used only for research and educational purposes. In preparation of this site and database, every effort has been made to offer the highest quality content. However, we make no warranty, expressed or implied, as to the accuracy of the information, in particular for the classification of the variants or its suitability for any specific purpose. Individuals, organisations and companies which use this database do so on the understanding that no liability whatsoever either direct or indirect shall rest upon the curators.
Analysis of missense variants
In order to help in interpretation of missense variants, protein alignments and Align GVGD [2,3] data are available below.
Access to protein alignments: These alignments have been elaborated using ortholog sequences or predicted ortholog sequences of the human proteins extracted from NCBI or Ensembl [4]. Only sequences which cover at least 80% of the human sequences have been considered. Alignments have been performed using ClustalW [5] (version 1.83.1). These alignments can be used for new variants as well as for existing ones. They are available in two different formats:
a text-formatted alignment, “human readable”, made using Edtaln, that contains accession numbers.
a clustal format (.aln) alignment that can be downloaded and displayed with an alignment editor such as the excellent Jalview [6].
alignments in text format:
cadherin 23 myosin VIIa protocadherin 15 sans harmonin usherin clarin 1
alignments in clustal format:
cadherin 23 myosin VIIa protocadherin 15 sans harmonin usherin clarin 1
Access to Align GVGD data: The preceding alignments have been submitted to the Align GVGD web program. This tool predicts the impact of an amino-acid substitution by combination of conservation and physico-chemical properties of residues. Variants are classified into 5 categories, which are, from the most deleterious to the lesser, ‘enriched deleterious 1 or 2’, ‘enriched neutral 2 or 1’. The fifth category is called ‘unclassified’.
Please note that Align GVGD data are only available for known missense variants*, and that these results are the output of a prediction software, and provided here on a “as is” basis, with no warranty as to their accuracy. We acknowledge Sean V. Tavtigian and the team of Align GVGD to allow us displaying these data.
cadherin 23 myosin VIIa protocadherin 15 sans harmonin usherin clarin 1
*last update: 2010-02-08
| -Brief overview of Usher syndrome:
Usher syndrome is an autosomal recessive disorder that associates both hearing and vision impairment. It is divided into three clinical subtypes (I, II and III) depending on the vestibular dysfunction and the degree and/or progression of hearing loss. Defects involve the cochlea and the retina. Vestibular dysfunction is observed in type I (the most severe form) and can be present in type III. More details can be found here 1 , 2. |
Both clinical and genetic heterogeneity are demonstrated (tables 1 and 2 below, [7]).
|
||||||||||||||||||||||||
Table 1: The different clinical types of Usher syndrome |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Table 2 : The different loci, genes and the encoded proteins
identified in Usher syndrome |
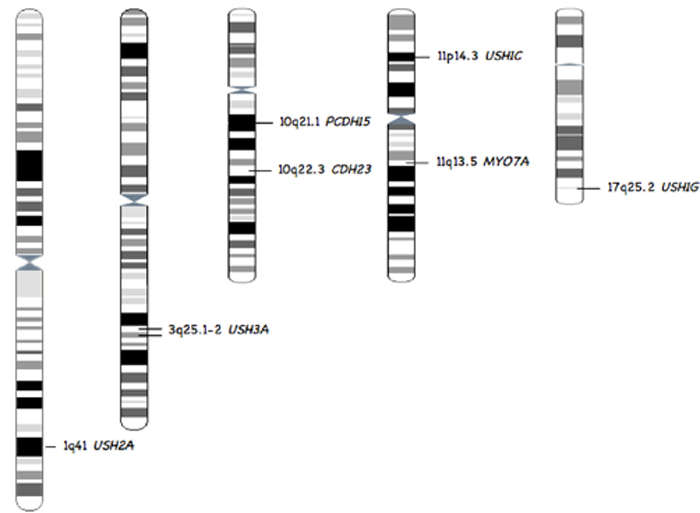 |
Figure 1: Chromosomal localisation of the 7 common Usher genes represented In the databases. Localisation extracted from genatlas.
 |
 |
References
[1] Beroud C, Hamroun D, Collod-Beroud G, Boileau C, Soussi T, Claustres M. UMD (Universal Mutation Database): 2005 update. Hum Mutat. 2005 Sep;26(3):184-91.
[2] Tavtigian SV, Deffenbaugh AM, Yin L, Judkins T, Scholl T, Samollow PB, de Silva D, Zharkikh A, Thomas A. Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J Med Genet. 2005 Jul 13.
[3] Mathe E, Olivier M, Kato S, Ishioka C, Hainaut P, Tavtigian SV. Computational approaches for predicting the biological effect of p53 missense mutations: a comparison of three sequence analysis based methods. Nucleic Acids Res. 2006 Mar 6;34(5):1317-25. Print 2006
[4] T. J. P. Hubbard, B. L. Aken, K. Beal1, B. Ballester1, M. Caccamo, Y. Chen, L. Clarke, G. Coates, F. Cunningham, T. Cutts, T. Down, S. C. Dyer, S. Fitzgerald, J. Fernandez-Banet, S. Graf, S. Haider, M. Hammond, J. Herrero, R. Holland, K. Howe, K. Howe, N. Johnson, A. Kahari, D. Keefe, F. Kokocinski, E. Kulesha, D. Lawson, I. Longden, C. Melsopp, K. Megy, P. Meidl, B. Overduin, A. Parker, A. Prlic, S. Rice, D. Rios, M. Schuster, I. Sealy, J. Severin, G. Slater, D. Smedley, G. Spudich, S. Trevanion, A. Vilella, J. Vogel, S. White, M. Wood, T. Cox, V. Curwen, R. Durbin, X. M. Fernandez-Suarez, P. Flicek, A. Kasprzyk, G. Proctor, S. Searle, J. Smith, A. Ureta-Vidal and E. Birney. Ensembl 2007. Nucleic Acids Res. 2007 Jan 1; Database issue.
[5] Thompson, J.D., Higgins, D.G. and Gibson, T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Research, 22:4673-4680.
[6] Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004 Feb 12;20(3):426-7. Epub 2004 Jan 22.
[7] Roux, AF. Molecular Diagnostic approaches to Usher syndrome. In ‘Molecular Genetic Analyses of Rare Diseases in 2007: Selected Examples’. 2007. Research Signpost. 103-120 ISBN: 81-308-0172-8.
[8] Petit C, Levilliers,J, Hardelin J.P. Molecular genetics of hearing loss. 2001. Annu Rev Genet 35:589-646.
[9] Kremer H, van Wijk E, Marker T, Wolfrum U, Roepman R. Usher syndrome: molecular links of pathogenesis, proteins and pathways. 2006. Hum Mol Genet 15 Spec No 2:R262-70.
[10] Reiners J, Nagel-Wolfrum K, Jurgens K, Marker T, Wolfrum U. Molecular basis of human Usher syndrome: deciphering the meshes of the Usher protein network provides insights into the pathomechanisms of the Usher disease. 2006. Exp Eye Res.
Pictures permissions:
“cochlea”: personal picture (AF Roux) of an Aztec stone rattlesnake, London British Museum.
“retina”: picture from Dr Hoover, used here with its personal authorization, original URL.
“chromosomes”: derived from Ensembl. Citation of the disclaimer: ‘Ensembl imposes no restrictions on access to, or use of, the data provided and the software used to analyse and present it. […] Some of the data and software included in the distribution may be subject to third-party constraints.’ We assume these pictures are not subject to third-party constraints.
 |
 |