The UMD-MLH1 mutations database
Mutation
c.IVS10-16del2 (c.885-16del2)
| Splice site type | Wild type sequence | CV | Mutant type sequence | CV | Variation (%) | |||||
| ctcttattttcctga |
| cttattttcctgaca |
| 62.1 % |
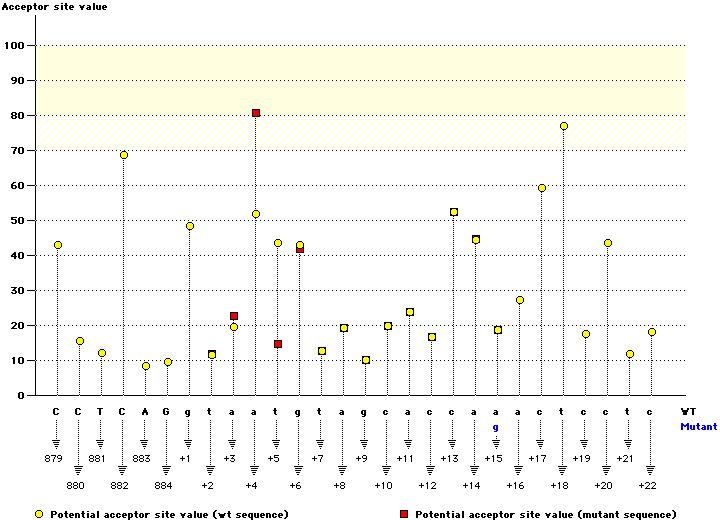 |
Data for this mutation
| Sample ID | MLH1 | MSH2 | MSH6 | MUTYH | APC |
|---|---|---|---|---|---|
| 7_4999 (05K255) 4160E0508 | c.IVS11-8T>A (c.1039-8T>A) c.IVS13+14G>A (c.1558+14G>A) | c.IVS1+9C>G (c.211+9C>G) | - | - | - |
| 8_41454_7091953_08-303290 | - | c.2242G>T (p.Asp748Tyr) | - | - | - |
| SO_-5463---G08440 | - | - | - | - | - |
| Analysis | Result | Date | Origin | PMID/dbSNP |
|---|---|---|---|---|
| Clinical phenotype | Sporadic colorectal cancer < 50 years old | 15/10/08 | SO (Marseille) | |
| MMR function in tumor cells | MLH1- | 18/10/04 | SO (Marseille) | |
| Ex vivo analysis | Splicing reporter minigene pCAS: normal splicing | 26/09/11 | 19 (INSERM U1079-Rouen) | |
| Clinical phenotype | Multiple colorectal cancers | 13/06/14 | 8 (Nancy) | |
| Co-occurrence | MSH2 : c.2242G>T; p.Asp748Tyr | 13/06/14 | 8 (Nancy) | |
| MMR function in tumor cells | MSI-H / MLH1+MSH2+MSH6+ | 13/06/14 | 8 (Nancy) |
| Biological significance | Date | Comment |
|---|---|---|
| Neutral | 26/09/11 | --- |