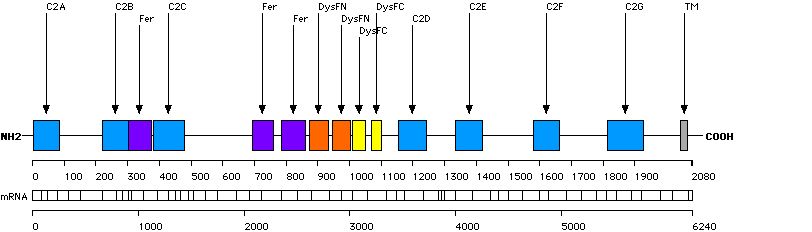
This function displays the distribution of small rearrangements in structural domains and key residues of the DYSF protein. Only mutations/variations found in probands are taken into account.
| Structural domain | Number of records |
|---|
| C2 Domain A [2-85] | 51 (2.6 %) |
| C2 Domain B [222-302] | 60 (3.1 %) |
| Ferlin family domain FerI [303-374] | 60 (3.1 %) |
| C2 Domain C [381-479] | 83 (4.3 %) |
| Ferlin family domain FerA [694-759] | 33 (1.7 %) |
| Ferlin family domain FerB [786-861] | 72 (3.7 %) |
| Outer DysF domain, N-terminal [874-933] | 85 (4.4 %) |
| Inner DysF domain, N-terminal [946-1002] | 122 (6.3 %) |
| Inner DysF domain, C-terminal [1011-1049] | 81 (4.2 %) |
| Outer DysF domain, C-terminal [1068-1101] | 11 (0.6 %) |
| C2 Domain D [1154-1244] | 42 (2.2 %) |
| C2 Domain E [1335-1421] | 108 (5.6 %) |
| C2 Domain F [1580-1663] | 136 (7.1 %) |
| C2 domain G [1813-1926] | 146 (7.6 %) |
| Transmembrane domain [2045-2067] | 4 (0.2 %) |
| Key residues (HCD) | Number of records |
|---|
| Calcium binding 16 [16-16] | 0 (0 %) |
| Calcium binding 21 [21-21] | 0 (0 %) |
| Calcium binding 71 [71-71] | 0 (0 %) |
| Calcium binding 73 [73-73] | 0 (0 %) |
| Calcium binding 288 [288-288] | 3 (0.2 %) |
| Calcium binding 411 [411-411] | 0 (0 %) |
| Calcium binding 417 [417-417] | 0 (0 %) |
| Calcium binding 465 [465-465] | 5 (0.3 %) |
| Calcium binding 467 [467-467] | 0 (0 %) |
| Calcium binding 473 [473-473] | 0 (0 %) |
| Calcium binding 1168 [1168-1168] | 1 (0.1 %) |
| Calcium binding 1174 [1174-1174] | 2 (0.1 %) |
| Calcium binding 1230 [1230-1230] | 0 (0 %) |
| Calcium binding 1232 [1232-1232] | 0 (0 %) |
| Calcium binding 1238 [1238-1238] | 0 (0 %) |
| Calcium binding 1348 [1348-1348] | 0 (0 %) |
| Calcium binding 1354 [1354-1354] | 2 (0.1 %) |
| Calcium binding 1407 [1407-1407] | 0 (0 %) |
| Calcium binding 1594 [1594-1594] | 0 (0 %) |
| Calcium binding 1600 [1600-1600] | 0 (0 %) |
| Calcium binding 1649 [1649-1649] | 0 (0 %) |
| Calcium binding 1651 [1651-1651] | 0 (0 %) |
| Calcium binding 1657 [1657-1657] | 1 (0.1 %) |
| Calcium binding 1849 [1849-1849] | 0 (0 %) |
| Calcium binding 1855 [1855-1855] | 2 (0.1 %) |
| Calcium binding 1912 [1912-1912] | 0 (0 %) |
| Calcium binding 1914 [1914-1914] | 0 (0 %) |
| Calcium binding 1920 [1920-1920] | 0 (0 %) |